Abstract
CD49d, the alpha-chain of the VLA-4 integrin heterodimer, is one of the strongest prognosticators in chronic lymphocytic leukemia (CLL), robustly identifying patients with dismal outcomes (Bulian et al, J Clin Oncol, 2014). While CD49d negative expression is stable due to a methylation-dependent regulation, its expression can be modulated by microenviromental stimuli (Zucchetto et al., Blood 2013; Benedetti et al, Leukemia, 2018). We demonstrated that ~20% of CLL shows a distinct bimodal pattern of CD49d expression with coexisting CD49d- and CD49d+ subpopulations, the latter increasing over time, especially after therapy, given the greater propensity to proliferate and segregate in tissue sites (Tissino et al, Blood 2020).
Here we investigated the evolutionary dynamics and epigenetic regulation of CD49d expression in CD49d bimodal CLL.
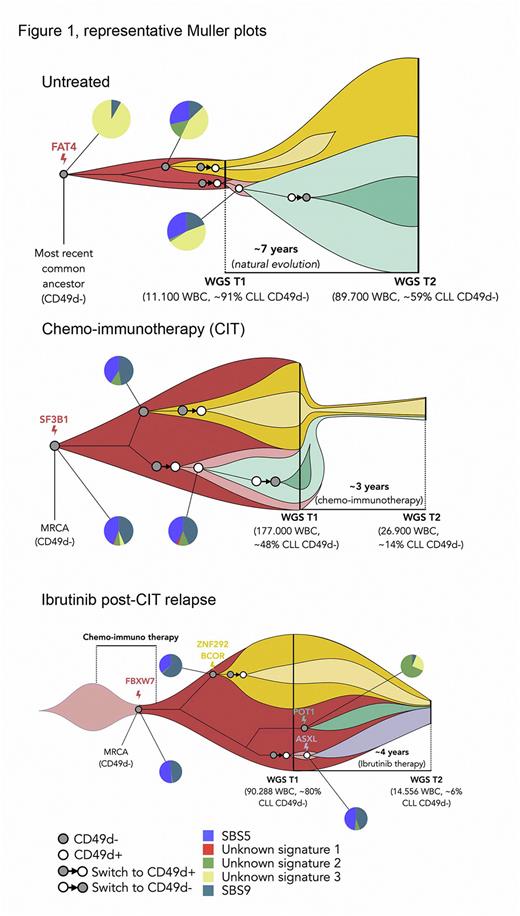
We collected two longitudinal samples (mean time interval, 4.3 years, range 2.0-7.0) from 6 CLL patients expressing bimodal CD49d, and performed separate 100X bulk whole genome sequencing (WGS) in sorted CD49d+ and CD49d- CLL cell (CD5+/CD19+) fractions in order to keep clonal dynamics of CD49d+ and CD49d- subpopulations distinguishable (n=24). Patients were either untreated (n=2) or treated with chemo-immunotherapy (CIT, n=2) or Ibrutinib post-CIT relapse (n=2) between the first and second sample. All patients had over time increase of the CD49d+ component.
WGS data was studied by state-of-the-art computational models for tumor subclonal deconvolution and somatic mutational signature profiling (Caravagna et al., Nat Genet, 2020; Lal et al., PLoS Comput Biol, 2021). Targeted analyses of DNA methylation (119 CpG islands) and chromatin accessibility (ATAC-seq) of the ITGA4/CD49d gene promoter were done by NGS with standard methods. RNA-seq was performed with the Illumina Stranded total RNA prep library kit. Multiparametric flow cytometry was used to discriminate the CLL proliferative (CD5high/CXCR4dim, PF) and resting (CXCR4high/CD5dim, RF) fractions, and to characterize tonsil B cell populations (CD19, CD3, CD27, IgM, IgD, 7-AAD).
From bulk WGS we observed, across all patients, strong signals of ongoing subclonal evolutionary dynamics. Somatic mutations highlighted clear switches among the CD49d+ and CD49d- components at both time points, strongly suggestive of CD49d rewiring transition of subclones with identical genetic origin (representative Muller plots in Figure 1).
In keeping with the driving role of CD49d in CLL: i) such transitions mainly occurred towards the upregulation of CD49d expression in CD49d- cells, although the opposite switch was also observed; ii) the most parsimonious explanation for the presence of subclones in both components was hypothesized to be a plastic pattern originating from a CD49d- cell rewiring CD49d expression to become CD49d+.
This raised the issue on CD49d expression in the putative CLL cell-of-origin (COO). In this regard, variable CD49d expression levels, from completely negative to strongly positive, were found in B cells at different maturation steps from normal B cell compartment of tonsil tissues (naïve, natural effector and memory B cells), confirming the suggestion that a CD49d- mature B cell could be the bona-fide COO for CLL with bimodal or negative CD49d expression.
The systematic evidence of CD49d plasticity found by WGS, prompted to investigate its epigenetic regulation. A higher degree of ITGA4 methylation was observed in the CD49d- fraction compared to the CD49d+ counterpart (p=0.004), confirming the methylation-driven epigenetic regulation of CD49d expression observed in other CLL settings (Zucchetto et al., Blood 2013). Consistently, progressively increasing chromatin accessibility of the ITGA4 promoter was observed by comparing CD49d- (n=3) vs CD49d bimodal (n=4) vs CD49d+ (n=11) CLL.
In keeping with Tissino et al. (Blood, 2020), the CD49d+ CLL component was enriched in cells recently egressed from lymph nodes belonging to the PF, rather than cells from the RF, mostly long-lived quiescent cells navigating the bloodstream (p=0.001). Accordingly, CD49d+ CLL cells overexpressed (p=0.0001) the transcription signature of the PF reported by Calissano et al (Mol Med, 2011), as well as the signature of CD49d+ CLL (Zucchetto et al., Cancer Res, 2009).
Overall, our WGS approach revealed that CD49d expression is plastic in CD49d bimodal CLL, such plasticity driven by epigenetic events.
Disclosures
Del Principe:Jansenn: Honoraria; Gilead Sciences: Honoraria; Incyte: Honoraria. Rossi:AstraZeneca: Consultancy, Honoraria, Other: Travel Support, Research Funding; Gilead: Other: honoraria, advisory board fees , Research Funding; Janssen: Consultancy, Honoraria, Other: Travel Support, Research Funding; AbbVie: Consultancy, Honoraria, Other: Travel Support, Research Funding; BeiGene: Consultancy, Honoraria, Other: Travel Support, Research Funding; MSD: Other: advisory board fees ; BMS: Consultancy, Honoraria, Other: Travel Support.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal